Developed to detect pathogens missed in wounds of soldiers, this technology was licensed to a company for development into a test for use by clinical laboratories
Diagnostic technology developed for rapid detection of pathogens in the wounds of soldiers has been licensed to a private company that intends to use it to create new medical laboratory tests. This new technology is capable of identifying thousands of bacteria and viruses in a single test.
Scientists at the Lawrence Livermore National Laboratory developed what is called the Lawrence Livermore Microbial Detection Array (LLMDA). Within 24 hours, this single test can detect multiple viruses and bacteria. The LLMDA technology has been licensed to St. Louis, Missouri-based MOgene LC, a supplier of DNA microarrays, according to a report published by UC Health.
New Microbial Detection Technology is Cheap, Fast and All Inclusive
LLMDA is designed to improve on two pathogen identification techniques: Polymerase chain reaction (PCR) analysis and DNA sequencing. PCR diagnostic techniques can process no more than 50 DNA signatures at one time, and the likelihood of discovering new species are low with PCR, noted a report published in LLNL’s Science and Technology Review.
Sequencing is a highly effective approach. It provides the most comprehensive information about both known and unknown pathogens. The drawback to sequencing, however, is that this process is costly and can take several days to produce results.
On the other hand, LLMDA is less expensive than sequencing and more inclusive than PCR analysis. Its developers say that LLMDA can detect and identify any bacterium or virus that has been previously sequenced.
The microbial detection array (MDA) process begins by purifying DNA or RNA from a blood or stool sample. The purified DNA or RNA is labeled with a fluorescent dye, then squirted onto the microarray which sits on top of an incubator heated to 42°C. The microarray contains nearly 400,000 probes arranged in a checkerboard pattern on a 2.5 by 7.5-centimeter glass slide.
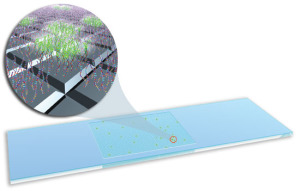
The DNA microarray (pictured above) contains nearly 400,000 probes and allows scientists to identify thousands of sequenced viruses and bacteria at the same time. Each square contains multiple strands of DNA or RNA. Squares are arrayed in orderly rows and columns on a 2.5- by 7.5-centimeter glass slide. (Rendering by Sabrina Fletcher, copyright by Lawrence Livermore National Laboratory.)
Scientists examine these probes with a fluorescent scanner and analysis software. The microarray’s checkerboard has several dozen squares for each of the thousands of organisms sequenced to date. That allows it to simultaneously examine multiple regions from each organism.
Nex-Gen Microbial Array Building More Detection Capacity
LLNL scientists are already building on the capacity of this microbial array. They are currently testing a next-generation LLMDA that contains 2.1 million probes representing about 178,000 sequences from 5,700 viruses and 785,000 sequences from thousands of bacteria. The new version also includes about 237,000 sequences from hundreds of fungi and about 202,000 sequences from 75 protozoa.
“We are designing the next-generation array to be the most comprehensive in the world,” stated Crystal Jaing, Ph.D., a biologist in Livermore’s Physical and Life Sciences Directorate who leads the LLMDA research. “Our goal is to develop a cost-effective technology that can detect both known and unknown pathogens within 24 hours,” she added.

Lawrence Livermore National Laboratory Biologist Crystal Jaing, Ph.D. (pictured above), holds up a slide containing the Lawrence Livermore Microbial Detection Array, which can identify thousands of known viruses and pathogens within 24 hours. (Photo copyright Lawrence Livermore National Laboratory.)
After designing a first-generation microarray several years ago, the LLNL bioinformatics team collaborated with researchers at University of California at San Francisco; University of Texas Medical Branch at Galveston; National Institute for Public Health and the Environment in Bilthoven, Netherlands; Statens Serum Institut in Copenhagen, Denmark; Imigene in St. Petersburg, Florida; and the U.S. Food and Drug Administration (FDA).
Interest by Military for Wound Care in Soldiers Moved Research Forward
Military medical professionals dealing with severe, complex infections in soldiers wounded in combat were interested in this concept. The result was a three-year study by researchers at LLNL and four other institutions—the Naval Medical Research Center and Walter Reed Army Institute of Research in Silver Spring, Maryland; the Uniformed Services University of Health Sciences in Bethesda, Maryland; and the University of California Davis in Sacramento.
The study goal was to develop technology that allows caregivers to more effectively diagnose, treat, and accelerate the healing and rehabilitation of soldiers injured in combat, noted a report published by Medical Xpress.
The study was published in the Journal of Clinical Microbiology in July 2014. Researchers evaluated 124 wound samples from 44 soldiers injured in Iraq and Afghanistan.
One key finding of this study was that certain bacteria, such as Pseudomonas species and Acinetobacter baumannii—which are common hospital-related infections—were associated with wounds that did not heal successfully. Bacteria often related to the gastrointestinal system, such as E. coli and Bacteroides species, were also often found in wounds that did heal successfully.
Innovative Microarray Technology Holds Promise for Civilian Uses
“Wound microbes are unique to each patient,” observed Nicholas Be, Ph.D., an LLNL biomedical scientist. He was the study’s lead author and was quoted in the Medical Xpress report.
“The use of advanced detection technologies could provide clinicians with valuable information during treatment,” noted Be. “In particular, information on the presence of specific bacteria that more significantly impact the success of the healing response could guide therapy and allow for more accurate prediction of outcome.”

Lab scientists Nicholas Be, Ph.D. (left), and Jonathan Allen, Ph.D., examine the Lawrence Livermore Microbial Detection Array (LLMDA), which can detect any virus or bacteria that has been previously sequenced within 24 hours. The LLMDA fits on a one-by-three-inch glass slide. (Photo copyright Lawrence Livermore National Laboratory.)
Jaing, who co-authored the study, foresees many civilian medical applications for the LLMDA technology. She said that it could be particularly useful in treating burns with large surface areas, people injured by trauma, or people with diabetic ulcers.
Slezak also noted that that the LLMDA technology could improve public health diagnostics because it can detect multiple bacteria and viruses in a single test. He pointed out, however, that clinical trials will be required to measure its efficacy in diagnosing human diseases and as a precursor to gaining FDA approval.
How long will it take for this diagnostic technology to find its way into clinical laboratories? Researchers did not offer an estimate. However, the fact that the technology has been licensed to MOgene LC is a sign of confidence that, with further development and good clinical study data, LLMDA technology might end up in medical laboratory tests following review and clearance by the FDA.
—By Patricia Kirk
Related Information:
Lawrence Livermore Microbial Detection Array (LLMDA)
New technology detects pathogens in soldiers’ wounds
Livermore Lab licenses microbial detection array advance
Livermore Lab’s microbial detection array detects plague in ancient human remains



